nCounter® FLEX Analysis System With Dx Enablement
Digital Molecular Barcoding Technology for Translational Research

The nCounter® FLEX Analysis System with optional Dx enablement is a platform optimized for translational research. The nCounter FLEX’s Dx enablement package allows the system to be run in either Diagnostic mode or Life Science mode depending on requirements. This is the only approved platform capable of running the Prosigna® Breast Cancer Prognostic Gene Signature Assay.
The nCounter® FLEX Analysis System is flexible platform for translation of oncology signatures from research to the clinic. This Platform is specifically designed clinical applications.
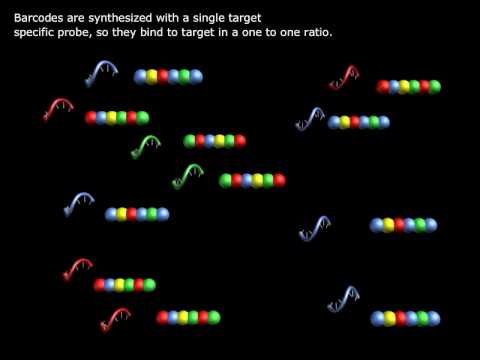
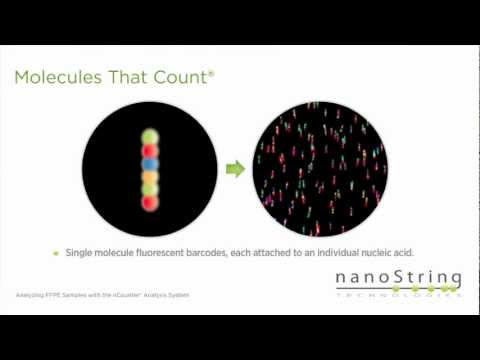
NanoString’s nCounter technology is molecular barcoding technology which digitally detects individual target molecules through the use of a unique probe pair for each target of interest. The probe pair consists of a color-coded Reporter probe, which carries the visible signal on its 5’ end, and a Capture probe, which carries a biotin moiety on the 3’ end. with a specific target. Each Reporter probe has six positions, and each position can be one of four colors. A large diversity of probes can be mixed together with the sample in a single well, with each Reporter probe interacting with a specific target. These target-probe complexes are then individually resolved and identified during data collection.
CLINIC-READY PLATFORM
The Prosigna® Breast Cancer Prognostic Gene Signature Assay has received 510(k) clearance for use with the nCounter Dx Analysis System with FLEX configuration. nCounter FLEX is manufactured under GMP guidelines and ISO 13485 to ensure quality and compliance with international standards.
TRANSLATIONAL SUITE
nCouner FLEX can operate in two distinct modes: Diagnostics and Life Sciences. In addition, an enterprise software package is included to control user access, automate data flow, and generate audit logs.
TUMOR ANALYSIS
Analyze gene expression without a cDNA synthesis step, enabling greater tolerance of RNA degradation and inhibitors associated with the formalin fixation process. Improve confidence when analyzing FFPE tissue such as tumors that can be matched to known patient outcomes.
DEVELOP TAILORED SIGNATURES
Reduce large gene sets (up to 800 targets) with a single nCounter CodeSet and single-tube multiplexing technology. Quickly reach a focused gene signature that will robustly identify tumor subtypes without the challenge of optimizing and maintaining components needed to support multiple RT-qPCR assays.
Simplify Analysis : No need for specialized Bioinformaticist. Results generated as direct counts and reported in a standard CSV file that can be imported into your favorite application or use the included nSolver Analysis Software for convenient data research analysis.
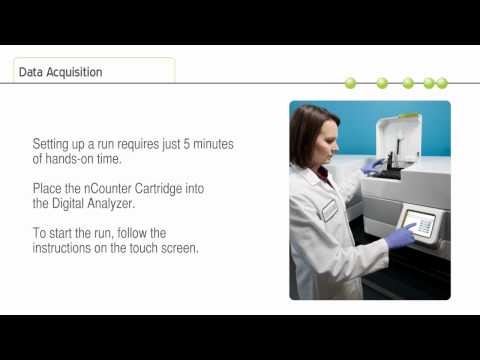
Expandable Throughput : Add a second Prep Station when needed and double your capacity to match a growing number of users. This configuration enables processing of up to 96 lanes per day and 384 samples with sample plexing. Random access enables users to pause data collection to load or remove additional cartridges without waiting for the run to finish.
|
Feature
|
Specifications RNA: 1 ng–50 ng DNA: 5 ng–300 ng Protein: 250 ng–2.5 µg Total RNA, cell lysates in GITC, FFPE-derived total RNA, and PAXgene-lysed whole blood Up to 30 µl 15 zeptomole spike-in control > 1.5-fold (if > 5 copies per cell) > 2-fold (if > 1 copy per cell) R² ≥ 0.95 6 x 10⁵ total counts 12 lanes per 2.5 hours 12 samples per 2.7 hours Assay dependent 50 base each for Capture and Reporter |
|
Hardware Specifications
|
18–28°C 30–80% (non-condensing) 2 100–240 VAC, 50–60 Hz 610 VA 67 x 89 x 63 cm 66 x 66 x 48 cm |
- The Prosigna® Breast Cancer Prognostic Gene Signature Assay
- Gene Expression Analysis - Analyze the expression levels of up to 800 genes simultaneously.
- miRNA Expression - Profile miRBase high confidence and published clinical miRNAs. Multiplexed target profiling of miRNA transcriptomes in a single reaction.
- miRGE Expression Analysis - Simultaneously profile miRNA and mRNA expression in a single reaction, Profile FFPE samples as well as other difficult sample types.
- Copy Number Variation Analysis - Profile up to 800 regions of the human genome in a single reaction, Better, faster results with less effort than PCR or arrays.
- lncRNA Expression Analysis- High precision, digital quantification of lncRNAs, Analyze up to 800 lncRNAs in a single reaction with no amplification.
- Leukemia Fusion Gene Analysis - Profile a comprehensive set of fusion genes in different leukemia subtypes.
- Single Cell Analysis - Obtain single cell sensitivity with reverse transcription and limited amplification.
- ChIP-String Assays - Skip the hassle of library prep and amplification when analyzing ChIP'd material.
- RNA:Protein Profiling - Measure RNA and protein simultaneously for true multi-omic data.
- DX analysis software only for Prosigna
- nSolver™ Analysis Software (nSolver™ 3.0)
- nCounter® Advanced Analysis Software